









This plasmid can be inserted into either bacterial or animal cells. Introducing DNA into bacterial cells can be done by
transformation (via uptake of naked DNA), conjugation (via cell-cell contact) or by transduction (via viral vector). Introducing
DNA into eukaryotic cells, such as animal cells, by physical or chemical means is called transfection. Several different
transfection techniques are available, such as calcium phosphate transfection, electroporation, microinjection and liposome
transfection. DNA can also be introduced into eukaryotic cells using viruses or bacteria as carriers, the latter is sometimes
called bactofection and in particular uses Agrobacterium tumefaciens. The plasmid may be integrated into the genome,
resulting in a stable transfection, or may remain independent of the genome, called transient transfection.
In either case, DNA coding for a protein of interest is now inside a cell, and the protein can now be expressed. A variety of
systems, such as inducible promoters and specific cell-signaling factors, are available to help express the protein of interest at
high levels. Large quantities of a protein can then be extracted from the bacterial or eukaryotic cell. The protein can be tested
for enzymatic activity under a variety of situations, the protein may be crystallized so its tertiary structure can be studied, or, in
the pharmaceutical industry, the activity of new drugs against the protein can be studied.
Polymerase chain reaction (PCR)
The polymerase chain reaction is an extremely versatile technique for copying DNA. In brief, PCR allows a single DNA
sequence to be copied (millions of times), or altered in predetermined ways. For example, PCR can be used to introduce
restriction enzyme sites, or to mutate (change) particular bases of DNA, the latter is a method referred to as "Quick change".
PCR can also be used to determine whether a particular DNA fragment is found in a cDNA library. PCR has many variations,
like reverse transcription PCR (RT-PCR) for amplification of RNA, and, more recently, real-time PCR (QPCR) which allow for
quantitative measurement of DNA or RNA molecules.
Real-time PCR
In molecular biology, real-time polymerase chain reaction, also called quantitative real time polymerase chain reaction
(Q-PCR/qPCR/qrt-PCR) or kinetic polymerase chain reaction (KPCR), is a laboratory technique based on the PCR, which is
used to amplify and simultaneously quantify a targeted DNA molecule. For one or more specific sequences in a DNA sample,
Real Time-PCR enables both detection and quantification. The quantity can be either an absolute number of copies or a
relative amount when normalized to DNA input or additional normalizing genes.
Expression cloning
One of the most basic techniques of molecular biology to study protein
function is expression cloning. In this technique, DNA coding for a protein of
interest is cloned (using PCR and/or restriction enzymes) into a plasmid
(known as an expression vector). A vector has 3 distinctive features: an
origin of replication, a multiple cloning site (MCS), and a selective marker
(usually antibiotic resistance). The origin of replication will have promoter
regions upstream the replication/transcription start site.
General overview of methods used in the lab
Macromolecule blotting and probing
The terms northern, western and eastern blotting are derived from what initially was a molecular biology joke that played on the
term Southern blotting, after the technique described by Edwin Southern for the hybridisation of blotted DNA. Patricia Thomas,
developer of the RNA blot which then became known as the northern blot actually didn't use the term.[2] Further combinations of
these techniques produced such terms as southwesterns (protein-DNA hybridizations), northwesterns (to detect protein-RNA
interactions) and farwesterns (protein-protein interactions), all of which are presently found in the literature.
Southern blotting
Named after its inventor, biologist Edwin Southern, the Southern blot is a method for probing for the presence of a specific DNA
sequence within a DNA sample. DNA samples before or after restriction enzyme digestion are separated by gel electrophoresis
and then transferred to a membrane by blotting via capillary action. The membrane is then exposed to a labeled DNA probe that
has a complement base sequence to the sequence on the DNA of interest. Most original protocols used radioactive labels,
however non-radioactive alternatives are now available. Southern blotting is less commonly used in laboratory science due to the
capacity of other techniques, such as PCR, to detect specific DNA sequences from DNA samples. These blots are still used for
some applications, however, such as measuring transgene copy number in transgenic mice, or in the engineering of gene
knockout embryonic stem cell lines.
Northern blotting
The northern blot is used to study the expression patterns of a specific type of RNA molecule as relative comparison among a set
of different samples of RNA. It is essentially a combination of denaturing RNA gel electrophoresis, and a blot. In this process RNA
is separated based on size and is then transferred to a membrane that is then probed with a labeled complement of a sequence of
interest. The results may be visualized through a variety of ways depending on the label used; however, most result in the revelation
of bands representing the sizes of the RNA detected in sample. The intensity of these bands is related to the amount of the target
RNA in the samples analyzed. The procedure is commonly used to study when and how much gene expression is occurring by
measuring how much of that RNA is present in different samples. It is one of the most basic tools for determining at what time, and
under what conditions, certain genes are expressed in living tissues.
Western blotting
Antibodies to most proteins can be created by injecting small amounts of the protein into an animal such as a mouse, rabbit,
sheep, or donkey (polyclonal antibodies) or produced in cell culture (monoclonal antibodies). These antibodies can be used for a
variety of analytical and preparative techniques.
The procedure follows the general principle of polymerase chain
reaction; its key feature is that the amplified DNA is detected as
the reaction progresses in real time. This is a new approach
compared to standard PCR, where the product of the reaction is
detected at its end. Two common methods for detection of
products in real-time PCR are: (1) non-specific fluorescent dyes
that intercalate with any double-stranded DNA, and (2)
sequence-specific DNA probes consisting of oligonucleotides
that are labeled with a fluorescent reporter which permits
detection only after hybridization of the probe with its
complementary DNA target.
Frequently, real-time PCR is combined with reverse transcription
to quantify messenger RNA and Non-coding RNA in cells or
tissues.
Gel electrophoresis
Gel electrophoresis is one of the principal tools of molecular biology. The
basic principle is that DNA, RNA, and proteins can all be separated by
means of an electric field. In agarose gel electrophoresis, DNA and RNA
can be separated on the basis of size by running the DNA through an
agarose gel. Proteins can be separated on the basis of size by using an
SDS-PAGE gel, or on the basis of size and their electric charge by using
what is known as a 2D gel electrophoresis.
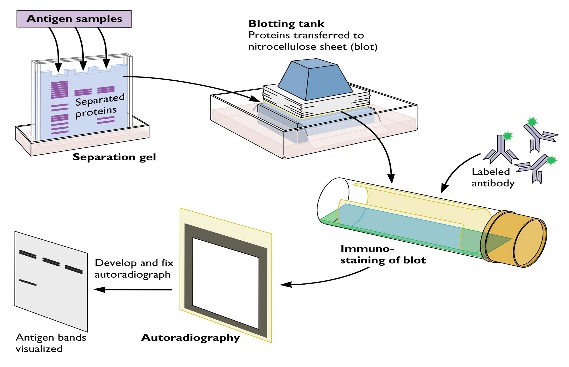
In western blotting, proteins are first
separated by size, in a thin gel sandwiched
between two glass plates in a technique
known as SDS-PAGE (sodium dodecyl
sulfate polyacrylamide gel electrophoresis).
The proteins in the gel are then transferred to
a PVDF, nitrocellulose, nylon or other support
membrane. This membrane can then be
probed with solutions of antibodies.
Antibodies that specifically bind to the protein
of interest can then be visualized by a variety
of techniques, including colored products,
chemiluminescence, or autoradiography.
Often, the antibodies are labeled with
enzymes. When a chemiluminescent
substrate is exposed to the enzyme it allows
detection. Using western blotting techniques
allows not only detection but also quantitative
analysis.
Text material above is adopted from Wikipedia for benefit of incomming students.




